
In the Lucent Laboratory, we study the physics of important biological processes on the molecular level using computer simulation. Most of our work involves simulating various characteristics of proteins including their assembly (folding), as well as their ability to interact with drugs (protein-ligand interactions) and their ability to act as catalysts (enzyme design).

Proteins constitute the fundamental structural and functional components of all life. They are nature’s nanomachines and they perform nearly all of the processes needed to keep you alive. They are made inside the cell by attaching one of twenty possible parts (amino acids) end to end in a growing chain (your DNA tells the cell the order in which to attach each part). Then, after the chain is formed, it somehow twists and turns and folds up into a functional shape (like molecular origami!). The mystery is how the protein knows how to fold itself up. How is the information needed to fold encoded in the sequence of parts? What are the most relevant physical principles needed to describe this? Why does folding sometimes go wrong and how does this cause diseases such as Alzheimer's disease, Parkinson's disease, cystic fibrosis, and many others? These are the questions that we as scientists aim to answer. Unfortunately, it is nearly impossible to follow the motions of all of the atoms in a protein quickly enough to see the protein fold. For this reason, we use computer simulations (mainly molecular dynamics which move the atoms of the protein according to Newton's second law) in which we input what we know about amino acids and the physics of small organic molecules and watch our simulations to observe how the proteins move (and when they fold and misfold). One of the most powerful and effective techniques for simulating protein folding involves using hundreds of thousands of individual computers working together to do small parts of a much larger molecular dynamics simulation. This approach is called distributed computing and is best known in the context of protein folding through the Folding@Home project.
In the Lucent Lab, we are interested in protein folding in vivo. This means that we are interested in simulating the things that make protein folding different inside a test tube (where we study proteins in the laboratory) compared to inside a cell (the place where proteins are actually doing their jobs). The environment inside the cell is much more crowded and complicated. Furthermore, special molecules called chaperonins are frequently needed to help proteins reach and maintain their functional shape. These chaperonins are basically big molecular barrels that swallow a misfolded protein and help it to fold. In the our lab, we use molecular dynamics simulation and distributed computing to figure out how proteins fold in crowded cytoplasm as well as inside the bellies of these large chaperonins.
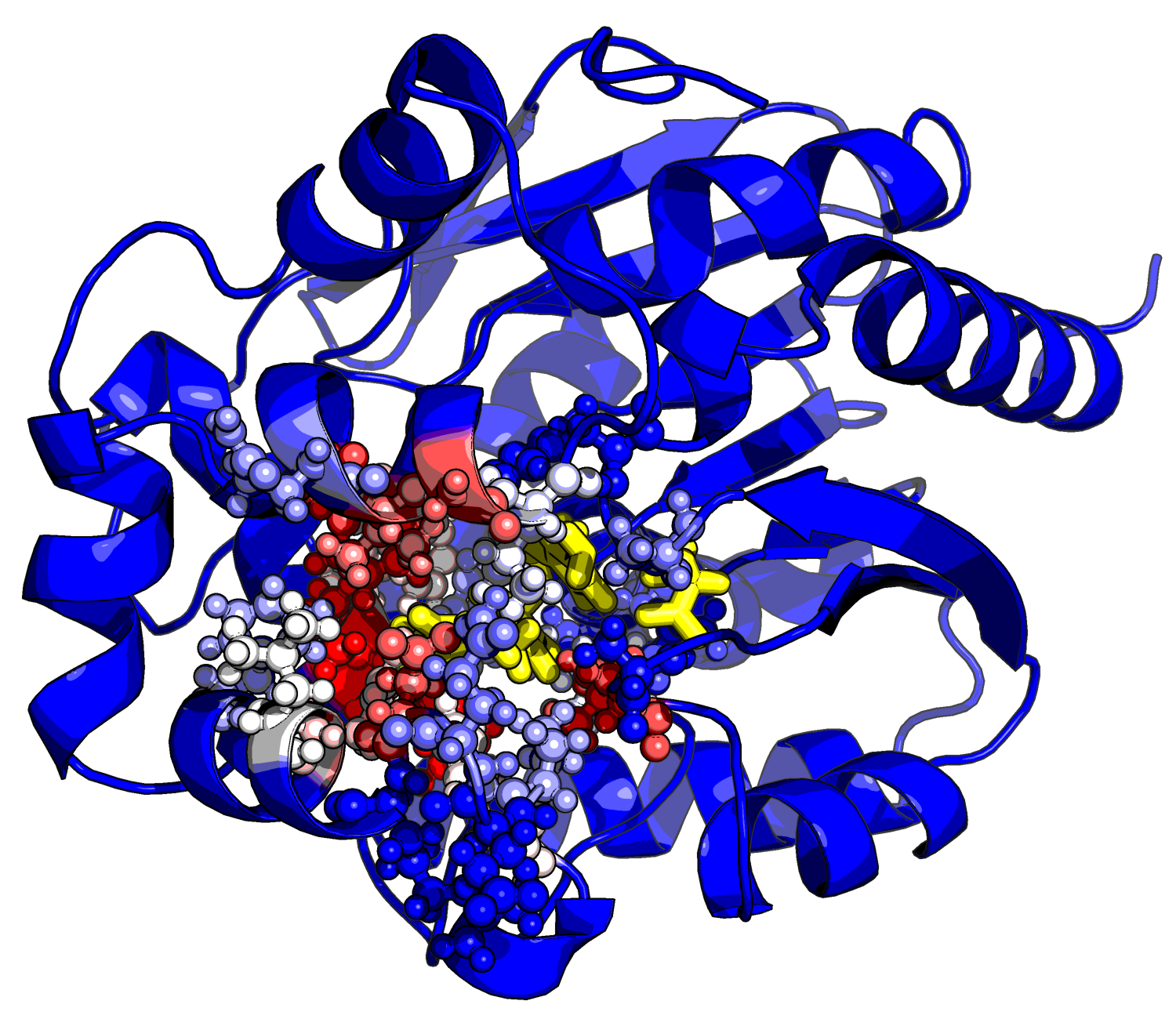
Once a protein is folded, one of the roles it can have is to act as a catalyst for a chemical reaction. Most chemical reactions necessary for life cannot occur inside a cell without a catalyst. A catalyst is something that speeds up a chemical reaction and does not itself get used up in the process (just like the catalytic converter in your car's exhaust system). Catalysts that work inside your cells are called enzymes. Scientists have been studying how enzymes work for a long time, but are only recently beginning to understand the process well enough to custom-build their own enzymes. New computational methods have allowed initial steps to be taken towards this goal. Despite the significant breakthrough represented by these methods, they generate a large number of false positive results and have not yet been able to produce enzymes with catalytic power on par with many of those found in nature. We believe that by leveraging the power of distributed computing, molecular dynamics, and quantum mechanics we will be able to rapidly search and validate an ensemble of rationally designed enzymes for activity. This method will ideally enable us to use a computer to design our custom enzymes and predict which ones will work best before we make them in the laboratory.
There is tremendous need for rationally designed enzymes in our modern society. As such, in addition to developing techniques for designing better enzymes in a computer, we are currently using these methods to design new enzymes to help with important problems. For example, we are in the process of making dechlorinase enzymes able to detoxify pesticide waste products such as beta-HCH, atrazine, and imidacloprid. We feel that rationally designed enzymes such as these could be very powerful tools for bioremediation.
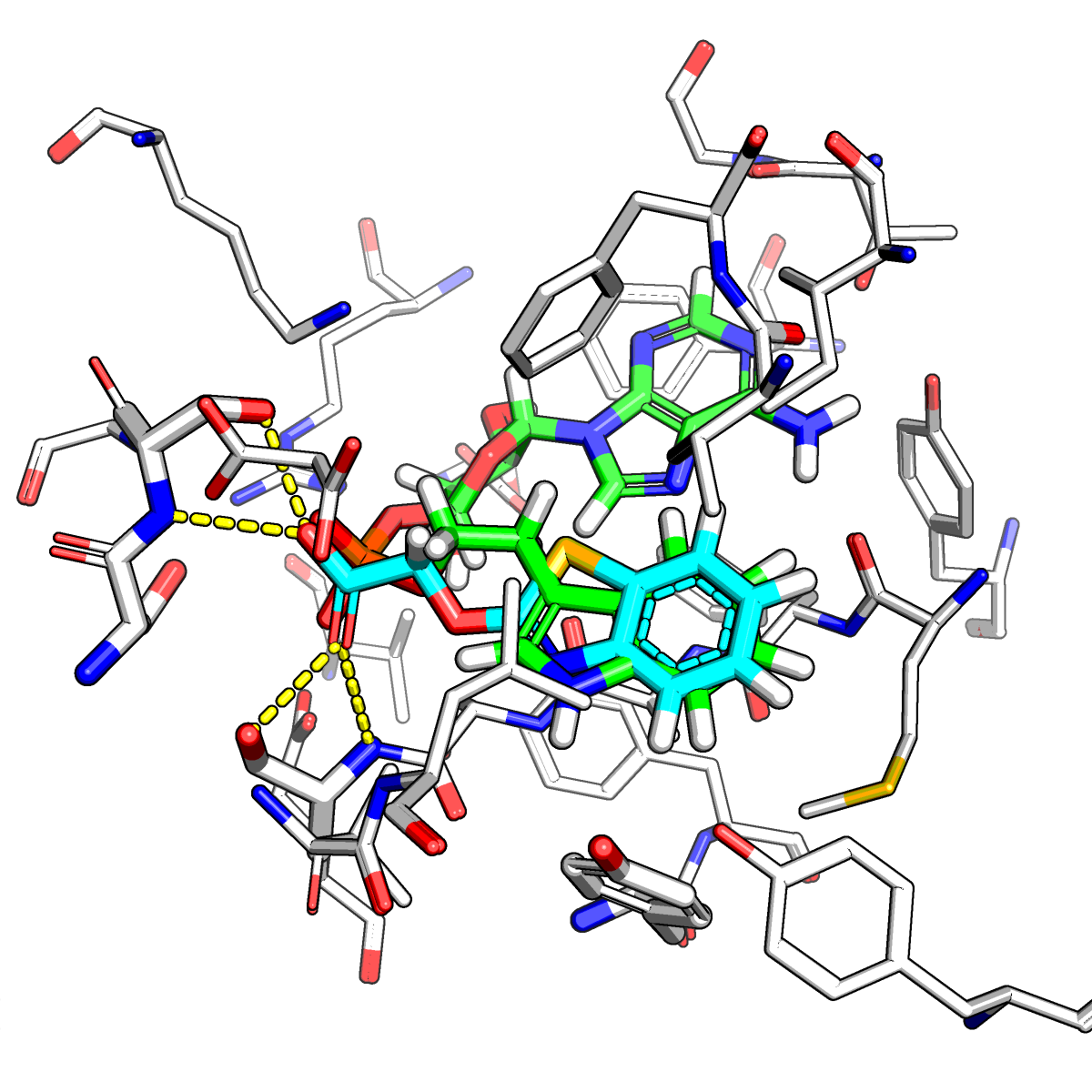
Just as proteins are large, diverse, and complicated molecules, they can interact with all sorts of smaller molecules. When one of these small molecules sticks to a protein, we typically call it a ligand and we call the protein a receptor (the terms ligand and receptor have more precise technical meanings, but they are colloquially used in this way). It turns out that most drugs fall into the class of some small molecule that sticks to a protein and somehow affects its function. Sometimes, drugs will bind to a protein to turn genes on or off, or to turn up or down the activity of an enzyme or signaling protein. Other times, drugs will bind tightly to part of a bacteria or virus in order to prevent it from working (thus killing it).
Despite the great number of drugs in existence, it is actually very hard to rationally design a drug and most existing drugs were discovered through serendipity and later refined for potency/toxicity/stability/etc. In order to accurately predict whether or not an arbitrary small molecule will bind to a specific protein (and in what orientation), we have to perform a number of simulations where we try to place the drug inside the protein. This type of calculation is referred to as docking. There are other types of simulations one can use to guess if a given small molecule would make a good drug, but in order to make mechanistic predictions that test our understanding of the relevant molecular structure and physics, some form of docking calculation must be performed (this is a part of structure-based drug design). In the Lucent lab, we aim to study how different proteins and ligands interact using a number of different simulation techniques (including docking, molecular dynamics, free energy perturbation, and Bayesian data analysis).